Making More Medicine Possible: MP
 When a drug works, it usually does by changing what a particular protein does.
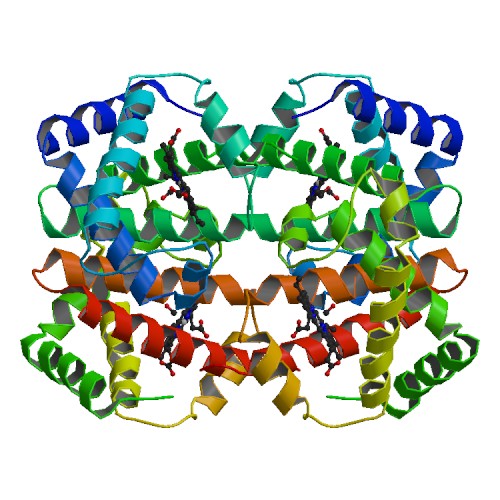
For example the drug Lipitor (pink) works because it stops the protein HMG-CoA Reductase
(blues) from creating the critical precursor to cholesterol. In order for a drug to change
what a particular protein does is needs to fit into that protein fairly specifically, like
a key into a lock. If you want to make a drug, it makes sense to want
to know the structure of the lock. But how much does it help really?
When a drug works, it usually does by changing what a particular protein does.
For example the drug Lipitor (pink) works because it stops the protein HMG-CoA Reductase
(blues) from creating the critical precursor to cholesterol. In order for a drug to change
what a particular protein does is needs to fit into that protein fairly specifically, like
a key into a lock. If you want to make a drug, it makes sense to want
to know the structure of the lock. But how much does it help really?
Large pharmaceutical companies may spend a billion dollars to develop a new drug. One-half of that money may be spent in pre-clinical drug development. If a pharmaceutical company has the three-dimensional structure of the protein the drug acts on, they can reduce those pre-clinical development costs by two-thirds. That can be as much as 333 million dollars. It does make sense to determine the three-dimensional structure of the target protein. And it is not merely about the money: drug development efforts informed by the drug target's structure access structure-based drug design. This method gave us many effective HIV protease inhibitors. Having the structure of the drug target may lead to the development of better drugs. Why do we not always do it?
Because the determination of a protein's three-dimensional structure can be quite difficult.
The determination of the structure of the protein that holds oxygen in the blood,
hemoglobin (above), took thirty years. 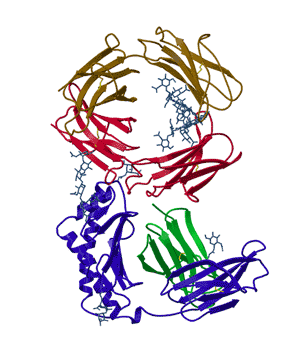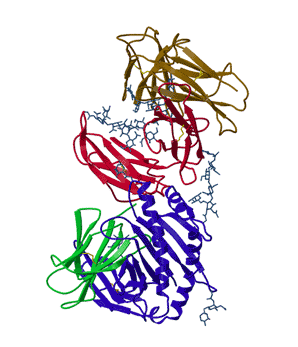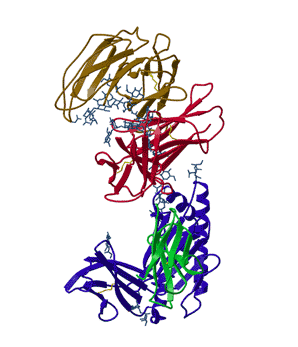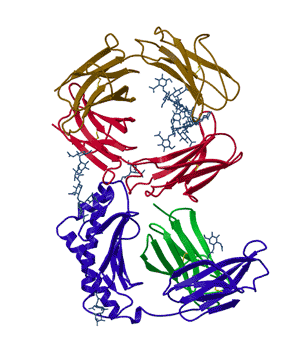
In his thesis work, Lance Martin determined the structure of the neonatal Fc receptor in complex with its ligand, the Fc portion of an IgG antibody to high resolution (above). Efforts to obtain that structure had gone on in his adviser's lab for a decade. Since that time, things have improved somewhat. There are now structural genomic efforts that are attempting to solve the structures of all the proteins in a given organism. They are making progress but majority of the proteins in a given genome still have no three-dimensional structures. Many of these proteins that are reluctant to reveal their structures are important drug targets. Why are they not feasible?

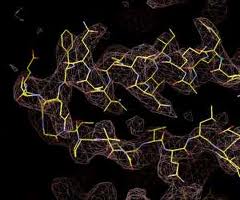
Most protein structure determination is accomplished by obtaining a crystal of the protein, bombarding that crystal with X-rays and using the diffraction pattern that results to constrain a model of the protein structure built by the protein crystallographer. The modern problem with this approach is obtaining protein crystals that diffract X-rays well. This process may take a day or it can take decades. There is no guarantee of success. Obtaining protein crystals usually requires a lot of protein. In order to acquire the three-dimensional structures of proteins that are refractory to the current methods, we are developing a method for acquiring that information that does not require protein crystals or even large amounts of protein.
Martin-Protean, LLC, was founded on April 8, 2007, by W. Lance Martin for the purpose of developing and bringing to market a technology for determining the three-dimensional structure of a protein without requiring protein crystals. Martin-Protean will supply pharmaceutical and biotechnology companies, as well as academic and national labs, the means to access the three-dimensional structure of proteins, protein-protein and protein-drug complexes.
In the process of developing this capability, Martin-Protean has developed a unique collection of protein modification reagents, peptide isolation techniques and peptide mass spectrometry capabilities. We can modify an intact protein and isolate the modified peptides from complex mixtures in 96-well format. In delivering peptide mass spectrometry services to clients we have also implemented these capabilities to analyze peptide and protein components of human fluids i.e., saliva, sweat, blood and urine. These fluids are important subjects for peptide analysis. The proteins and their quantities in these fluids are different between individuals, change over time, may be biomarkers for disease, and are not well-described by analyses informed by nucleic acids.
For details regarding our currently available products and services please see our protein structure or human fluid analysis pages. For recent events at Martin Protean, please consult our News.